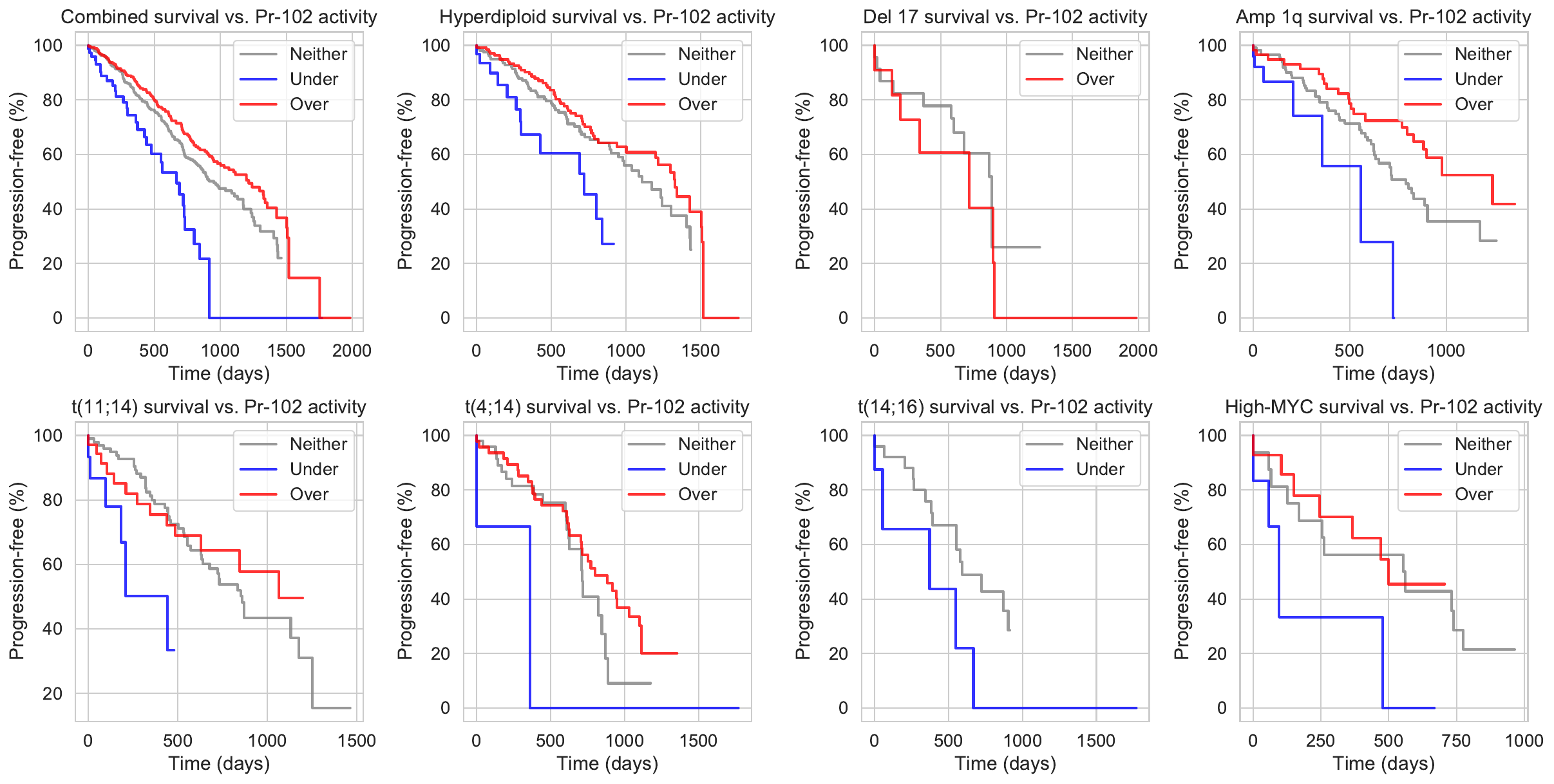
| t(11;14) | down-regulates | TP53 | activates | R-367 | -0.661782 | 11 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DCHS1 | down-regulates | ELF2 | represses | R-985 | -0.875385 | 8 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| FAT3 | down-regulates | ELF2 | represses | R-985 | -0.875385 | 8 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| NEB | down-regulates | BCL6 | represses | R-553 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| SYNE1 | down-regulates | BCL6 | represses | R-553 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del17 | down-regulates | BCL6 | represses | R-553 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(14;16)_cytogenetic | down-regulates | BCL6 | represses | R-553 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DCHS1 | down-regulates | ELF3 | represses | R-2155 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del17 | down-regulates | ELF3 | represses | R-2155 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del1p | up-regulates | ELF3 | represses | R-2155 | -0.963705 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| MAX | up-regulates | TCF3 | activates | R-2062 | -1.17031 | 20 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| RNASeq_MAFA_Call | down-regulates | TCF3 | activates | R-2062 | -1.17031 | 20 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(11;14) | down-regulates | NR2F6 | activates | R-1380 | -1.33754 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DCHS1 | up-regulates | E2F2 | activates | R-2947 | -1.79304 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DNAH5 | up-regulates | E2F2 | activates | R-2947 | -1.79304 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| FAT3 | up-regulates | E2F2 | activates | R-2947 | -1.79304 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| MAX | up-regulates | E2F2 | activates | R-2947 | -1.79304 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| PTPRD | up-regulates | E2F2 | activates | R-2947 | -1.79304 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(11;14) | down-regulates | ARID3A | activates | R-1355 | -2.23573 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(14;16) | up-regulates | ZNF410 | represses | R-1937 | -2.27804 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| amp1q | up-regulates | ZNF410 | represses | R-1937 | -2.27804 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del13 | up-regulates | ZNF410 | represses | R-1937 | -2.27804 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del1p | up-regulates | ZNF410 | represses | R-1937 | -2.27804 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(4;14) | up-regulates | XBP1 | activates | R-3060 | -2.38036 | 8 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del1p | down-regulates | XBP1 | activates | R-3060 | -2.38036 | 8 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DIS3 | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| RNASeq_MAFB_Call | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(14;16) | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| TRAF3 | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| TTN | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| amp1q | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del13 | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del1p | up-regulates | ELK4 | represses | R-769 | -3.00912 | 7 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| FGFR3 | up-regulates | CREB3L2 | activates | R-522 | -3.41802 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| RNASeq_FGFR3_Call | up-regulates | CREB3L2 | activates | R-522 | -3.41802 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| MYC | down-regulates | CREB3L2 | activates | R-522 | -3.41802 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(4;14) | up-regulates | CREB3L2 | activates | R-522 | -3.41802 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| TTN | down-regulates | CREB3L2 | activates | R-522 | -3.41802 | 6 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| DIS3 | up-regulates | NRF1 | represses | R-2706 | -4.26518 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(11;14) | up-regulates | NRF1 | represses | R-2706 | -4.26518 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| t(14;16) | up-regulates | NRF1 | represses | R-2706 | -4.26518 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| amp1q | up-regulates | NRF1 | represses | R-2706 | -4.26518 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |
| del13 | up-regulates | NRF1 | represses | R-2706 | -4.26518 | 10 | Pr-102 Genes (10)CCR4, KPNA4, NANOS3, NHLRC1, HIST3H2BB, DDX42, SH3D21, ZNF345, PINX1, CCPG1 Regulons (13)R-367, R-985, R-553, R-2155, R-2062, R-1380, R-2947, R-1355, R-1937, R-3060, R-769, R-522, R-2706 |