Mechanistic Inference of Node-Edge Relationships (MINER) pipeline
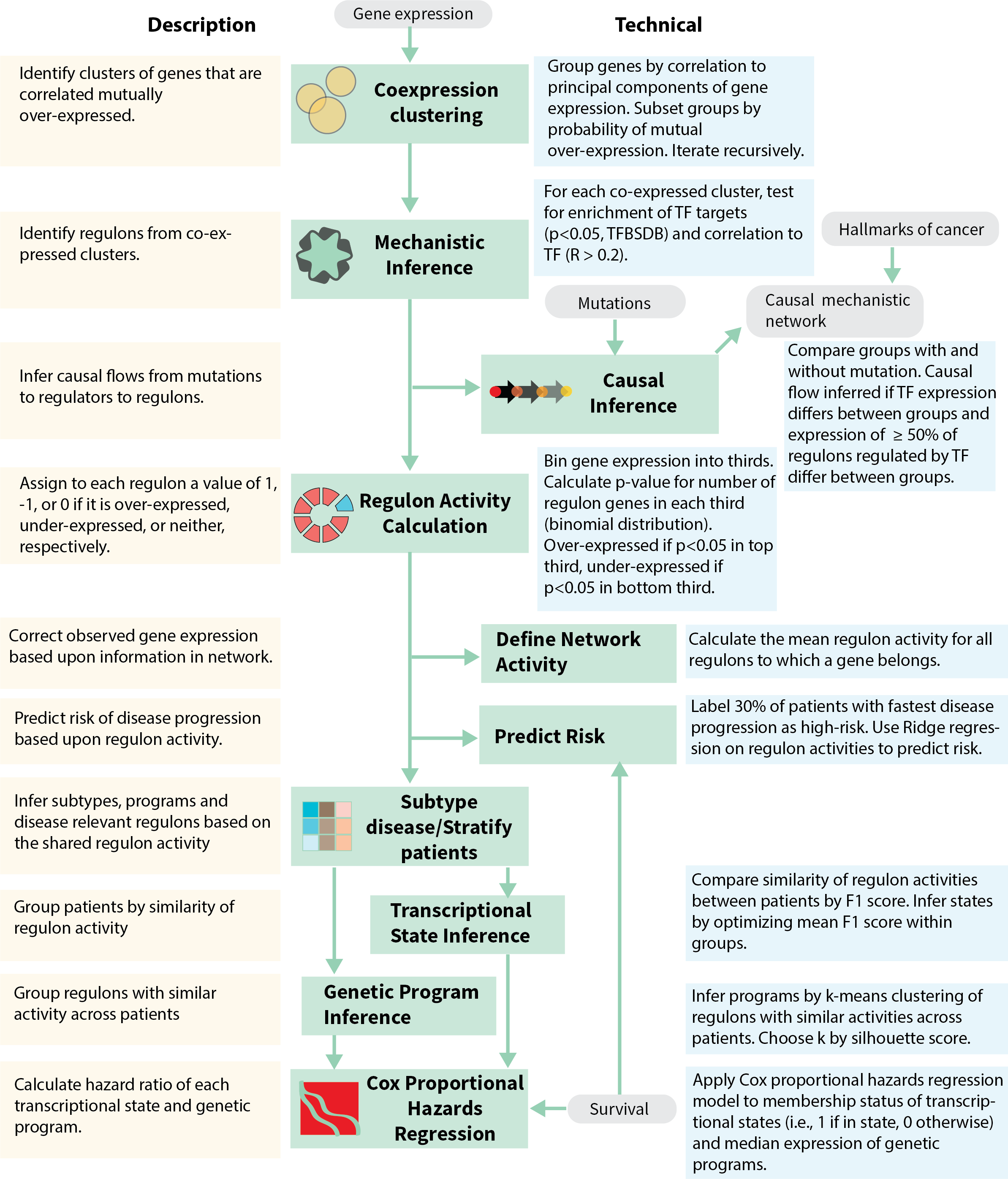
We developed the MINER pipeline to infer transcriptional regulatory networks from gene expression data and apply them to the characterization and prediction of phenotypes. MINER builds upon our previous work with the SYstems Genetics Network AnaLysis (SYGNAL) pipeline insofar as it enables the same core functionalities of mechanistic and causal inference, but does so with a new suite of algorithms that enable new applications in the network-based prediction of clinical outcomes (Fig. 1). Inference of the transcriptional regulatory network (TRN) begins by clustering gene expression data into coherent sets of genes that share a binding site for a transcription factor or miRNA according to gold-standard binding-site database information (e.g., Transcription Factor Binding Site Database – TFBSDB). By default, the cluster of genes and the corresponding regulator must also be correlated (or anticorrelated) to one another, however, this restriction can be lifted if it proves too stringent, for example in single-cell analysis. The combination of a coherently expressed set of genes and the associated regulator whose binding site they share represent discrete units, called regulons, from which the TRN is assembled. Once the regulons have been discovered, a novel causal inference algorithm (see Methods) identifies statistically significant links between putative causal events (somatic mutations, chromosomal translocations, etc.) and the activity levels of regulators and co-regulated gene sets. Once a MINER TRN has been inferred from the data of a patient cohort, new samples can be analyzed to uncover the disease-relevant modules that are over- or under-active in an individual patient.
Source Code
Source code is available at https://github.com/baliga-lab/miner2/
Documentation
Documentation is available at https://baliga-lab.github.io/miner2/

